Laboratory of In silico design
1. Members
| Project Leader | Suyong Re |
|---|---|
| Postdoctoral Research Associate | Yi-an Chen, NISHIMA Wataru |
| Trainee | Firoozeh Piroozmand |
| Administrative Assistant | TOJO Mitsuyo |
2. Background and objectives
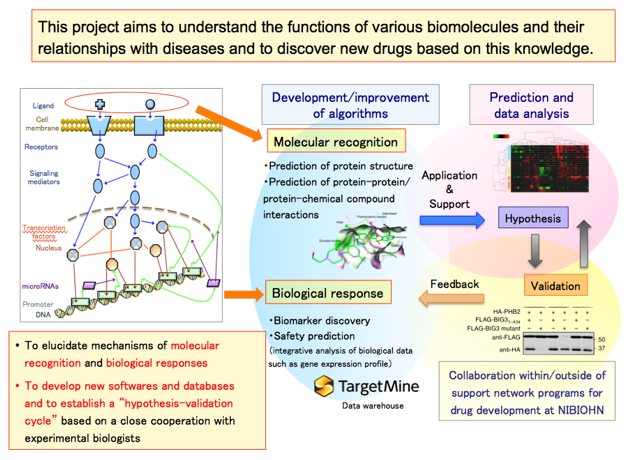
Our research aims to understand the functions and reactions of biological molecules and their relationships with diseases in an integrated system, and to discover new drugs based on this knowledge. Our goal is to improve the identification of drug targets by systems biology approaches, and to achieve effective selection/design of drug candidates by predicting their efficacy, pharmacokinetics and toxicity.
3. Overview of our research
Our main goal is to elucidate mechanisms of “molecular recognition” and “biological responses”. We develop our own software and databases, present new hypotheses to experimental biologists and receive feedback to iteratively improve our hypothesis, establishing a “hypothesis-verification cycle”.
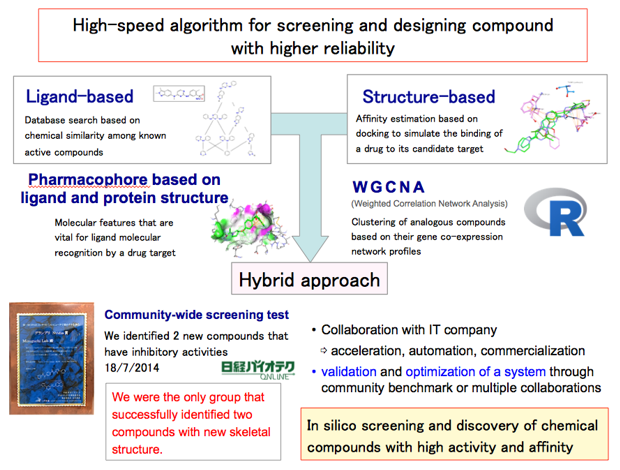
3-1. A case study in molecular recognition 1
We have developed a novel virtual screening protocol by combining ligand-based and structure-based approaches. In a community-wide blind test held in 2014, we were the only group that successfully identified two new inhibitory compounds with a novel scaffold for human c-Yes kinase.
References:
- Prathipati P, Mizuguchi K, Integration of Ligand and Structure Based Approaches for CSAR-2014, J. Chem. Inf. Model, 2015.
- Prathipati P, Mizuguchi K, Systems Biology Approaches to a Rational Drug Discovery Paradigm, Current Topics in Medicinal Chemistry, 16(9):1009-1025, 2015.
- Chiba S, Ikeda K, Ishida T, Gromiha M, Taguchi Y, Iwadate M, Umeyama H, Hsin K, Kitano H, Yamamoto K, Sugaya N, Kato K, Okuno T, Chikenji G, Mochizuki M, Yasuo N,Yoshino R, Yanagisawa K, Ban T T, Teramoto R, Ramakrishnan C, Thangakani A. M, Velmurugan D, Prathipati P, Ito J, Tsuchiya Y, Mizuguchi K, Honma T, Hirokawa T, Akiyama Y, Sekijima M, Identification of potential inhibitors based on compound proposal contest: Tyrosine-protein kinase Yes as a target, Scientific Reports, 5:17209, 2015.
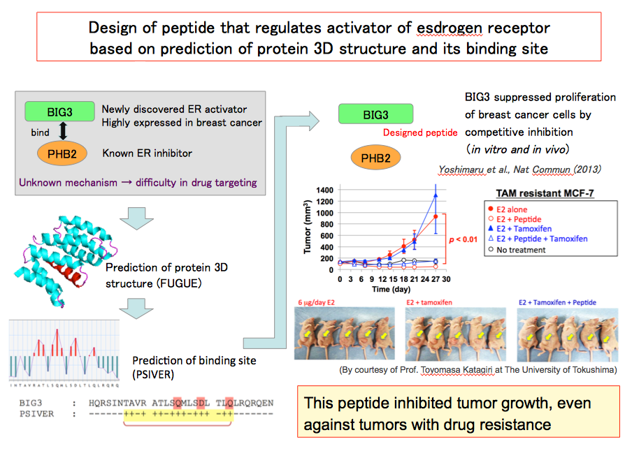
3-2. A case study in molecular recognition 2
The structural or functional information on the target is not always available and it can hinder an experimental investigation. To overcome this situation, we have developed methods for predicting protein structures or the amino acid residues that would interact with another protein from amino acid sequence information alone. We applied these methods to BIG3, a novel regulator of the estrogen receptor alpha, and demonstrated that a synthetic peptide mimicking the predicted interaction site suppressed the growth of tumor cells both in vitro and in vivo.
References:
- Murakami Y, Mizuguchi K. Applying the Naive Bayes classifier with kernel density estimation to the prediction of protein-protein interaction sites. Bioinformatics, 26:1841-1848, 2010.
- Yoshimaru T, Komatsu M, Matsuo T, Chen YA, Murakami Y, Mizuguchi K, Mizohata E, Inoue T, Akiyama M, Yamaguchi R, Imoto S, Miyano S, Miyoshi Y, Sasa M, Nakamura Y, Katagiri T. Targeting BIG3-PHB2 interaction to overcome tamoxifen resistance in breast cancer cells. Nat Commun, 4:2443, 2013.
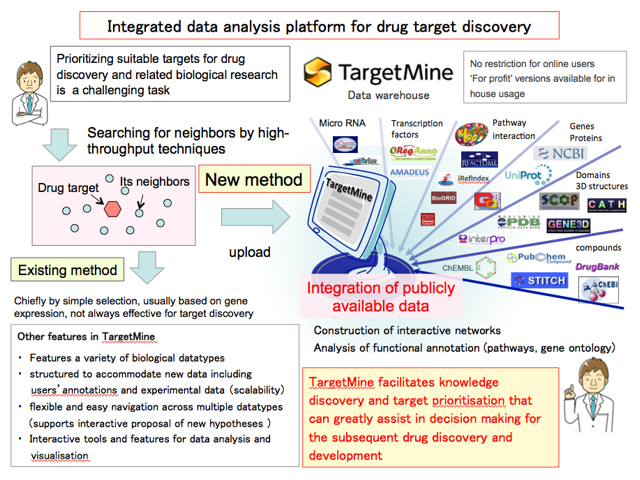
3-3. A case study in biological responses 1
Prioritising suitable targets is a key objective of drug discovery and related biological research, but it remains a demanding task. To achieve this, we have developed “TargetMine” an integrated data analysis platform that features many publicly available biological data types and analytical tools to analyse them.. TargetMine enables the users to discover new hypotheses related to biological phenomenon under study select promising candidate genes, pathways and networks for further investigation that that can greatly assist subsequent drug discovery and development.
We have also developed Toxygates, an integrative data analysis platform that seamlessly links chemical compound-induced gene expression profiles with pathways, protein-compound interactions and homologous associations over the semantic web. Toxygates enables the users to quickly and comprehensively assess compound toxicity and predict adverse drug responses
References:
- Chen YA, Tripathi L.P, Mizuguchi K, TargetMine, an integrated data warehouse for candidate gene prioritisation and target discovery, PLoS One, 8;6(3):e17844, 2011.
- Tripathi LP, Kambara H, Chen YA, Nishimura Y, Moriishi K, Okamoto T, Morita E, Abe T, Mori Y, Matsuura Y, Mizuguchi K. Understanding the biological context of NS5A-host interactions in HCV infection: a network-based approach. J Proteome Res, 12:2537-2551, 2013.
- Chen YA, Tripathi LP, Dessailly BH, Nystrom-Persson J, Ahmad S, Mizuguchi K. Integrated pathway clusters with coherent biological themes for target prioritisation. PLoS One, 9:e99030 , 2014.
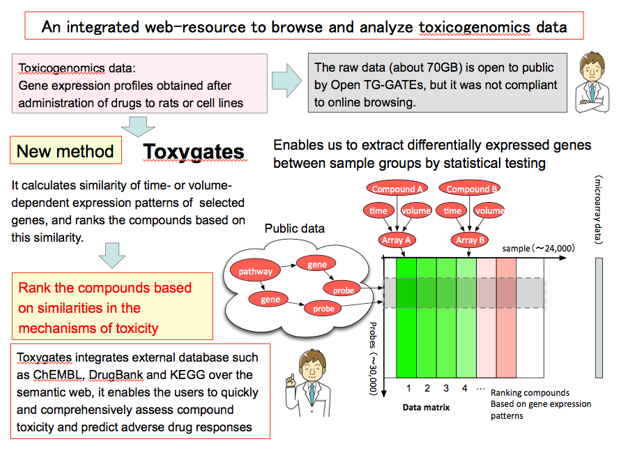
Reference:
- Nystrom-Persson J, Igarashi Y, Ito M, Morita M, Nakatsu N, Yamada H, Mizuguchi K. Toxygates: interactive toxicity analysis on a hybrid microarray and linked data platform. Bioinformatics, 29:3080-3086, 2013.
3-4. Example of a development of software infrastructure for drug designing
To prioritise suitable drug candidates, it is necessary to establish an integrated platform for in silico analysis that can predict ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) of chemical compounds with high accuracy. We are designing an ADMET prediction system that can leverage chemical structural information to develop QSAR (Quantitative Structure-Activity Relationship) models that can accurately predict predict physical parameters and help select suitable drug candidates in early stage drug developmet.
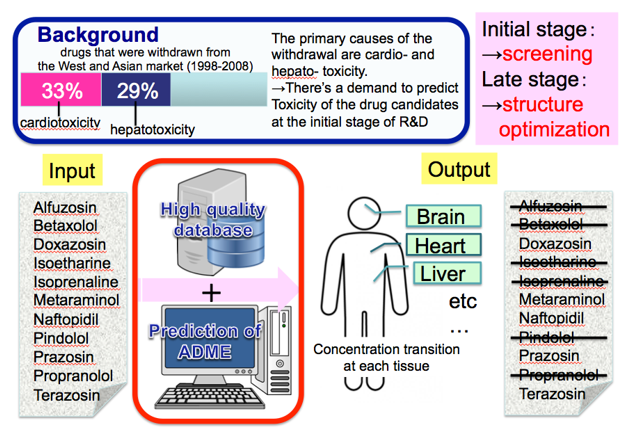
Other publications:
- Takemura N, Kawasaki T, Kunisawa J, Sato S, Lamichhane A, Kobiyama K, Aoshi T, Ito J, Mizuguchi K, Karuppuchamy T, Matsunaga K, Miyatake S, Mori N, Tsujimura T, Satoh T, Kumagai Y, Kawai T, Standley DM, Ishii KJ, Kiyono H, Akira S, Uematsu S. Blockade of TLR3 protects mice from lethal radiation-induced gastrointestinal syndrome. Nat Commun, 5:3492, 2014.
- Camargo LM, Zhang XD, Loerch P, Caceres RM, Marine SD, Uva P, Ferrer M, de Rinaldis E, Stone DJ, Majercak J, Ray WJ, Chen Y-A, Shearman MS, Mizuguchi K. Pathway-Based Analysis of Genome-Wide siRNA Screens Reveals the Regulatory Landscape of App Processing, PLoS One, 10(6):e0129641, 2015.
